Prediction of potential Protein Domains Using RNA-Seq Read-Mapping and Assembly Methods for Nipponbare, HiFi, and Nanopore Rice Accessions
InterProScan85 was used to predict potentialProteinDomains. For calculating gene-expression levels, low-quality RNA-seq reads were first removed using fastp86 (v.0.23.0) with parameters ‘-l 30’. The genome was mapped by Salmon87 against the index of decoysequences, using parameters ‘-l A –validateMappings-gcBias’. Finally, gene-expression levels were quantified by counting the number of reads mapping to each transcript and calculating the transcripts per million (TPM) values.
We implemented both read-mapping-based and assembly-based approaches to identify SNPs using Nipponbare as the reference genome. The alignment results of reads were used to call SNPs through Longshot 104. The parameters for the HiFi genomes were set to 3:10:50 and for the nanopore genomes were set to 3:10:50. The assembly-based calling process involves aligning each contig to a reference genome using a program such as thenu cmer program and refining the alignments to one-to-one matches. The show-snpp program was used to identify the genes, all from the MUMMER package. We only considered the SNPs detected by both methodologies in order to minimize false positives.
In 133 wild rice accessions and 129 cultivated rice accessions, thergawful90 was used to make a prediction. Four major families were identified and classified by the presence of RLKs, NLs, NBSs, and SM-CC genes in the family. The modules of the MCscan93 and the rcvi.para.Catalog were used to identify tandems, since they tend to cluster together. If the numbers were 1, 2 and more, they were categorized as singletons, pairs and clusters.
The method for constructing col linear blocks for each accession was used in the MCscan (Python version) pipechan. Tandem genes were integrated using the ‘mcscan’ tool of the ‘jcvi.compara.synteny’ module with the parameter ‘–mergetandem’. Then, all of the collinear blocks for each accession with all others were joined to a matrix using the ‘join’ tool of the ‘jcvi.formats.base’ module. A custom script was used to create a comprehensive RGA matrix, which was created by merging, sorting and deduplicating all collinear matrices.
To find the telomere sequence in each of the chromosomes, a custom script was used to look for the reverse complement of the seven bases.
In a previous study37, the dynamics of the LTR family were determined by comparing family size and the ratio of solo-to-intact LTRs within each family between two groups. Families showing significant differences in both sizes and the solo-to-intact LTR ratio were categorized as ‘removal families’. We separated the cases into two categories:amplification families and family size. On the other hand, if there was a notable change in the solo-to-intact LTR ratio without a corresponding shift in family size, these were classified as ‘balanced families’. Families that did not exhibit a significant difference in either dimension were termed ‘drifting families’. The dynamics of Or-IIIa- larger Gypsy families can be broken down into two parts, one for solo elements and another for intact elements. Then, the solo-to-intact ratio was calculated by dividing the number of solo elements by the number of intact elements within the Gypsy families. Finally, we applied Student’s t-tests to compare the family size and solo-to-intact ratio between Or-IIIa and japonica groups, with P < 0.01 as the cut-off for significance.
To detect changes, we aligned the pseudo-chromosome to the reference genome across the 149 genomes. Per the classifications provided by SyRI, INV variants were categorized as inversions, in comparison with the Nipponbare reference. Both Trans and INVTR were labeled as translocations. To detect small indels, we extracted INS and DEL variants consisting of fewer than 30 bp.
The pan-genome was transformed into a haplotype graph with the alignments of the 40 haplotype genomes to the DM reference using minimap2 (ref. 73). Genomic variants were identified using SyRI68. Specifically, the reference genome coordinates were binned into non-overlapping 100-kb windows. Haplotypes in each window were clustered together if the edit distance between their SNP profiles was less than 10% of the number of SNPs (that is, for any two haplotype instances assigned to the same cluster, they show less than 10 SNP differences per 100 SNPs to each other). The haplotype graph shows how each cluster of haplotypes is represented. The nodes in adjacent windows were connected by edges if they were linked in any of the contributing haplotypes. For each node, we identified marker k-mers (k = 51) that were (1) in all the contributing haplotypes, (2) in regions being in synteny with DM and (3) unique to that node.
To assemble the genomes of 16 O. sativa, 129 O. rufipogon and a total of 145 accessions, we applied the Minigraph-Cactus pipeline107 to them. The primary pangenome graph was created using minigraph108 and capturing the SVs within the input assembly. minigraph108 was used to re map these assemblies onto the primary graph. The mapping results were then used as the input for Cactus109 (v.2.2.1), which facilitated the generation of the final graphs. We defined the graph size as the total length of all nodes, and nodes that were not included in the reference genome (non-ref) were defined as novel sequences. We need to call for 142 accessions from our study, and 407 newly mapped samples from another study. The variations were then called using DeepVariants110 (v.1.6.1) with the NGS model, and all individual variants were merged using GLnexus111 (v. 1.4.1-0-g68e25e5).
We performed an all-versus-all comparison of the amino acid sequences of protein-coding genes using DIAMOND114 (v.2.0.15). These genes were from 149 genome assemblies and 31 assemblies (excluding the same species NIP and WSSM) from a cultivated rice pangenome16. The alignment results were then input into OrthoFinder115 (v.2.5.4) to find orthogroups and orthologues. Using 844 identified single-copy orthologues, we constructed a gene-based maximum-likelihood phylogenetic tree using IQ-TREE96 (v.2.2.0.3) with 1,000 bootstraps.
We used MSMC2. To infer the population separation history. Our analysis began with the preparation of a negative mask file for the coding region of IRGSP-1.0 (MSU7.0) and a mappability mask file using seqbility (http://lh3lh3.users.sourceforge.net/snpable.shtml) (v.20091110) and makeMappabilityMask.py. The phased SNP sites with uniquely mapped reads and mean coverage depths greater than threefold were acquired using Longshot104 (v.0.4.1) and the high-quality regions of each genome were acquired using the filtered results of show-snps from MUMmer66 (v.4.0.0beta2). The MSMC2 input files were built using a script called thegenerate_multihetsep.py. Because O. rufipogon naturally uses both cross-pollination and self-pollination, we followed an established approach of constructing pseudodiploids, which has been widely used in similar studies of inbreeding species such as Caenorhabditis123, Arabidopsis thaliana124, soybean125 and African wild rice126,127. We randomly selected four samples from each population and treated each sample as a single haplotype. We then used the haplotypes we had from the same population to make pseudodiploids. The population split inference focused on 2 individuals (4 haplotypes) per group, calculating median population split times based on 50 random combinations for each comparative analysis. A mutation rate of 8.09 × 10−9 per site per generation128 and a generation time of one year were applied to estimate demographic history.
The nucleotide diversity (π) of each group and the fixation index (FST) between different groups were both estimated using VCFTools119 (v.0.1.16) with a window size of 100 kb and a step size of 10 kb. Plot_MultiPop.pl was used to plot the genome-wide decay pattern for each group and the parameters were included in the PopLDdecay package. PLINK116 used the options of the ‘–genome’ and the ‘–genome-full’. The ggplot2 package is part of the R package.
We used the qp3Pop program to perform an F3-admixture test to detect potential admixture events of the form. The expected F3 statistic would yield a non- negative mean if the null hypothesis is true. A negative mean of the F3 statistics would suggest that genes from source 1 and source 2 are related to the population. A Z-score between 3 and 5 is indicative of significant ancestry in population C.
Using a four-taxon model (((P1, P2), P3), PO), we calculated the D-statistic to perform the ABBA–BABA test, using the script calculate_abba_baba.r (https://github.com/palc/tutorials-1/tree/master/analysis_of_introgression_with_snp_data/src). With O. longistaminata designated as the outgroup, our analysis revealed a significantly positive D-statistic (P < 0.01), suggesting introgression between P3 and P2. To delve deeper into introgression segments between indica and aus from japonica, we computed the fd statistic across the genome in 100-kb sliding windows with a step size of 10 kb, using the script ABBABABAwindows.py from genomics_general toolkit (https://github.com/simonhmartin/genomics_general). The minimum number of SNPs per window was set to 250, and the minimum proportion of samples genotyped per site was set to 0.4. The fd 0 and 1 values are converted to zero. Finally, to assess the congruence of introgression regions between indica and aus from japonica, we catalogued the putative introgression segments within the top 10, 30 and 50 100-kb windows.
We analyzed all possible combinations of varieties to determine the genetics of the two groups. We looked at the same windows as those with a similarity of more than 99%. The similarity index for each 10-kb window was calculated using the following formula:
The geographical records of all wild rice in the study were obtained by collecting field samples. Approximate latitude and longitude information on their distribution ranges was used for spatial mapping, and this can be found in Supplementary Table 2. The base map layers were derived from a public-domain Natural Earth dataset, used in the distribution map.
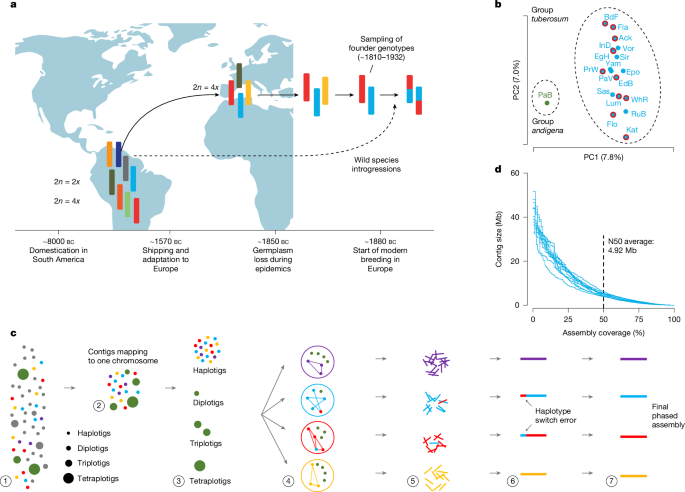
The leaves from 19 different countries were analysed and the genetic material was obtained from each of them with the Nucleo Mag plant kit. The libraries were sent to BGI, Hongkong (China) on dry ice, where they were sequenced on a DNBseq platform (Supplementary Table 2). Principal component analysis was used to analyze these sequencing samples to select the ten most diverse cultivars for pan-genome construction (Supplementary Methods). Blinding and randomization were not used.
The syntenic analysis at the whole genome level between the 4 representative genomes in O. rufipogon and O. longistaminata. Nested blocks were identified and visualized using the mumplot program.
We performed a comparative analysis between the pseudo-chromosomes obtained with the ALLMAPS method and the chromosomes constructed using Hi-C technology. At first, we aligned the genomes using the nucmer program from MUMmer66. The block was identified through a filter using the parameters ‘-m’. In addition, we compared alignments between the two chromosomes with identified synteny blocks and structural rearrangements.
High-molecular-weight (HMW) DNA was isolated from 1.5 g of material with a NucleoBond HMW DNA kit (Macherey Nagel). Quality was measured with a FEMTOpulse device, and quantity was measured by fluorometry. According to the manual, the library was prepared using the SMRTbell Express template. Size distribution was again controlled by FEMTOpulse (Agilent). Size-selected libraries were sequenced on a Sequel II device at Max Planck Genome-centre Cologne (MP-GC) with Binding kit 2.0 and Sequel II Sequencing Kit 2.0 for 30 h. (Read statistics are provided in Supplementary Table 2).
The Pan-Genome of F-Wild Potatoes (Supplementary Table 2): A Deep Variant Calling Analysis based on Mosdepth Alignment
The potato cultivars (Supplementary Table 2) were clonally propagated and grown on Murashige–Skoog medium for 3–4 weeks at Max Planck Institute for Plant Breeding Research (MPIPZ, Germany). Plantlets were transferred to soil in 7 × 7-cm2 pots and grown in a Percival growth chamber for 2–3 weeks. The plants were transferred to pots and grown until they flowered. The plants were grown in very cold conditions, 22 C.
Using short reads of a query genome, marker k-mers were extracted using Jellyfish51 (v.2.2.10). The probability of zero, one, two, three or four haplotypes was estimated with a mixture model. A process of expectation maximization was used to determine if a sample was zero, one, two, three, or four copies, or if all the samples were equal. Nodes with a non-zero copy number were then heuristically connected to form pseudo-contigs.
A single haplotype was used to create the pan-genome. Further haplotypes were iteratively incorporated using alignments against the haplotypes that were already included in the pan-genome using minigraph (v.0.20-r55966)27 with parameters ‘-cxggs -t 20’. A model, y = a1 × x/(x + a2) + a3, fitting the increasing pattern of the pan-genome size was constructed, for which the parameters were optimized using the BFGS method in R 4.3.0 (ref. 83).
The whole-genome reads of 20 wild potato species were aligned using minimap2 (v.2.20-r1061). Variant calling was performed using DeepVariant (v.1.4.0)74. The variant were merged into a single dataset. To evaluate potential introgressions, Read Depth was calculated across the genomes using Mosdepth.
Haplotype-specific sequences of each cultivar were aligned to the reference genome double monoploid (DM) 1-3 516 R44 using nucmer3 (v.3.1)67,70. Structural variations and syntenic regions are referred to as SNPs. The distribution of structural variation across the genome was determined using Msyd (v.1.0) (https://github.com/schneebergerlab/msyd).
For each chromosome, each of the 40 haplotype-specific sequences were aligned to each other using nucmer3 (v.3.1)67 with options ‘–maxmatch -c 100 -l 80 -b 500’. The files were processed with options which included the option of show-coords and the option of calling single-. The chromosome-level comparisons were visualized with a modified version of plotsr69.